About proGenomes v2.1
proGenomes v2 (Nucleic Acids Res doi: 10.1093/nar/gkz1002) is an update of proGenomes (Nucleic Acids Res doi: 10.1093/nar/gkw989).
The most recent update to version 2.1 provides 84,096 consistently annotated bacterial and archaeal genomes. In this update, genomes flagged for contamination were removed to provide a higher quality dataset. Taxonomic annotations are provided as species clusters (Mende et al., Nature Methods, 2013) and as NCBI taxonomy. Functional annotations of over 280 million genes are provided as eggNOG orthologous groups (Huerta-Cepas et al., NAR, 2016) and for antibiotic resistance and virulence.
We further provide a set of 40 universal, single-copy genes for each of the genomes (Cicarelli et al., Science. 2006; Sorek et al., Science, 2007) to support phylogenetic studies of the genomes.
Additionally, 12,226 representative genomes covering all species clusters are available for direct download and these can be used for the annotation of metagenomics datasets, large scale phylogenetics and other comparative approaches. Within representative genomes, we also provide habitat specific sets.
Start exploring proGenomes by searching for a taxonomic group or species cluster or an individual genome.
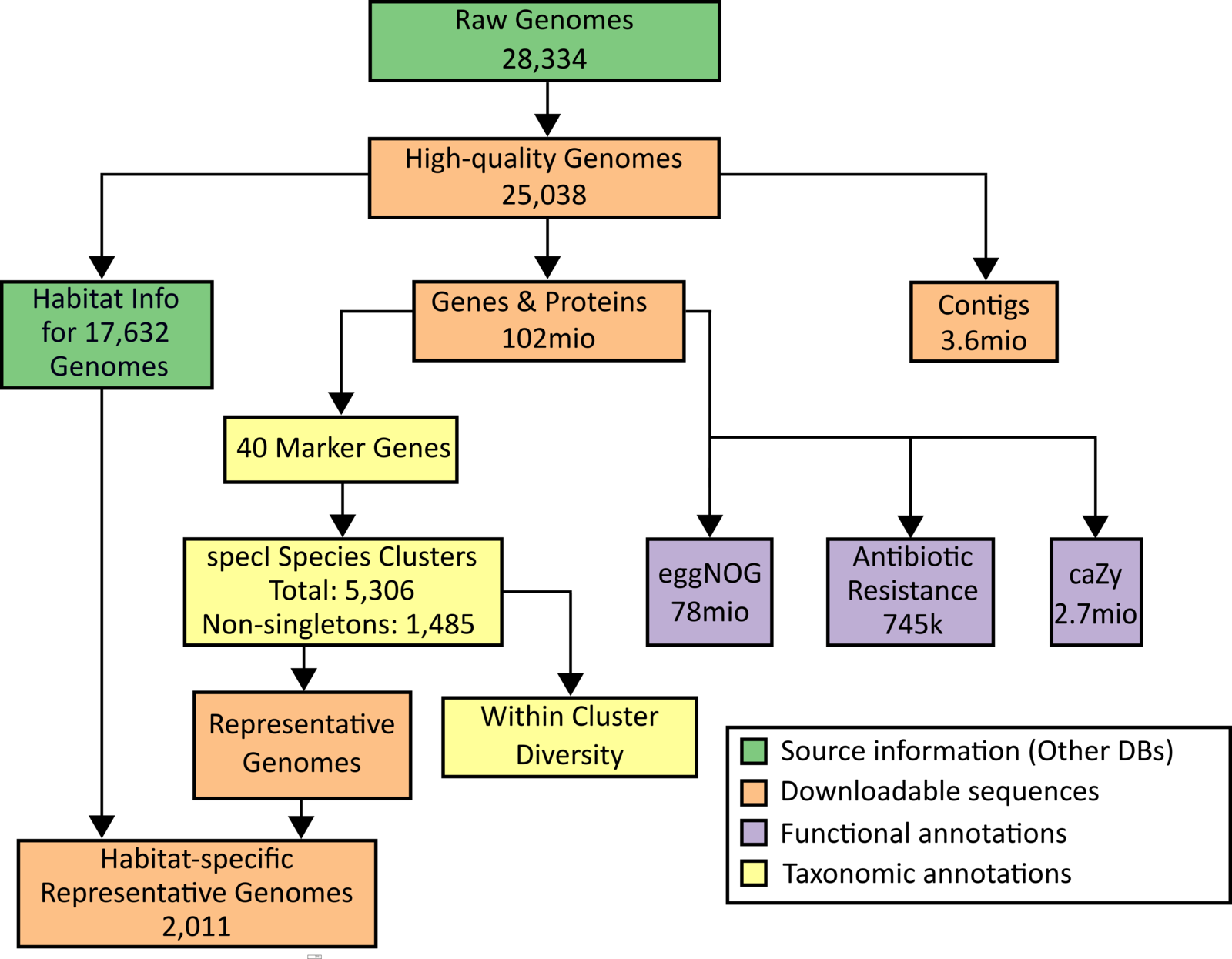
Workflow to generate the underlying data of the database
Previous versions
Previous version of the database is available at the following address: http://progenomes1.embl.de/
Contact
proGenomes 2 is free for academic and non-commercial use. For commercial use or customized versions, please contact biobyte solutions GmbH.
We hope you find the database user-friendly and easy to use. However, if you encounter any problems or have questions, please